Bromodomain Inhibitors – an exciting new target in cancer research?
One of the interesting things about basic cancer research is that new targets emerge all the time, offering fresh opportunities for developing novel therapeutics in the quest for clinical improvement. While you see many companies chasing the same well established targets, often generating me-toos, sometimes serendipity favours the bold and the brave, as we recently saw with Pfizer’s development of crizotinib for ALK+ lung cancer.
So what’s new on the R&D front?
Bromodomain inhibition is a novel cancer target and one that I am looking forward to learning more about at forthcoming annual meeting of the American Association for Cancer Research (AACR) in Washington DC.
The plenary session on Monday April 8 on Epigenetic Targets in Cancer includes a presentation by James Bradner (Dana-Farber Cancer Institute) on Bromodomain Inhibition in Cancer.
What are BET bromodomains?
Although there are 47 bromodomain proteins, a subset of four proteins are associated specifically with the BET bromodomain, including a terminal (T) node:
- BRD2
- BRD3
- BRD4
- BRDT
These bromodomains are acetyl-lysine binding pockets that target bromodomain-containing proteins to histones and thereby affect chromatin structure and function. The binding of BET protein bromodomains to chromatin regulates gene expression. Whenever histones are involved, epigenetics are never far behind.
Thus in simple terms, it is now believed that targeting the binding of bromodomain and extra-terminal [BET] proteins to chromatin, it may be possible to regulate gene expression, and in particular, the transcription of key oncogenes such as MYC, which can lead to arresting of cell-cycle progression and apoptosis (programmed cell death). In plain English, this means that cancer cells are selectively killed.
Some of the groups original preclinical work in this area was published in Nature a couple of years ago – it was ground breaking work because generally protein-protein interactions such as MYC are considered very difficult targets to drug, unlike tyrosine protein kinases (TKIs), which are more accessible. More recent work by Lin et al., (2012) in Cell elaborated on the significance of low and high MYC levels.
Since then, the research has moved into the translational and clinical space. Small molecule inhibition of BET is a drug development target of Cambridge, MA based Constellation Pharmaceuticals, who announced last September that they partnered with The Leukemia & Lymphoma Society to develop their novel BET for the treatment of hematologic malignancies.
Highlighting the significance of this work is the announcement in January this year (see Fierce Biotech’s piece for more details) is a $95M deal with Genentech, which includes a buyout option. I thought this was a smart move at the time, given the importance and solidity of the basic research findings. The venture funds invested in privately-held Constellation Pharmaceuticals clearly have an exit strategy in mind.
I am hoping that Dr Bradner’s AACR plenary presentation will discuss Constellation’s drug development pipeline.
What makes BET bromodomain inhibitors of even more interest as a potential target is the possibility that there may be biomarkers that will identify those patients most likely to respond to therapy. For those of you interested in a basic understanding of biomarkers, you can read more on Biotech Strategy Blog for an overview.
Preclinical research published by the group in the March 2013 edition of Cancer Discovery highlights how a genetic biomarker could be used to identify those cancer patients likely to respond to a new class of cancer drugs, BET Bromodomain Inhibitors.
MYC is overexpressed in many cancers but until recently, has largely been ignored as an ‘undruggable target’. A rare malignant childhood cancer known as neuroblastoma, for example, is associated with the amplification of the MYCN gene and generally considered to be difficult to treat unless it is caught early and surgical resection is feasible. New work published in Cancer Discovery by Puissant et al., gives hope that therapeutic targeting with bromodain inhibitors might be a feasible strategy to pursue.
Recent research published in 2011 in the Proceedings of the National Academy of Science (PNAS) by Jennifer Mertz et al., at Constellation Pharmaceuticals showed that you could target MYC dependence in cancer by inhibiting BET bromodomains. They showed that:
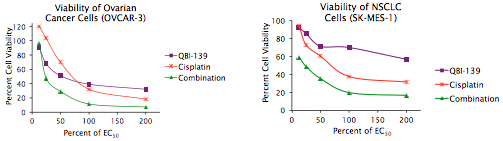
“Small molecule inhibition of BET bromodomains leads to selective killing of tumor cells across a broad range of hematologic malignancies and in subsets of solid tumors.”
Fast forward to the work published in Cancer Discovery. Using high-throughput screening the researchers at Dana Farber found that amplification of the MYCN gene in neuroblastomas was sensitive to BET bromodomain inhibitors. Using cell lines with MYCN amplification and a mouse model of neuroblastoma, they showed that BET bromodomain inhibitors prolonged survival and had anti-tumor effects.
The Cancer Discovery abstract describes the significance of this work:
“Biomarkers of response to small-molecule inhibitors of BET bromodomains, a new com- pound class with promising anticancer activity, have been lacking. Here, we reveal MYCN amplification as a strong genetic predictor of sensitivity to BET bromodomain inhibitors, show a mechanistic rationale for this finding, and provide a translational framework for clinical trial development of BET bromodomain inhibitors for pediatric patients with MYCN-amplified neuroblastoma.”
What does all this data mean?
It has long been known that MYC protein is likely oncogenic in some tumour types, including some hematologic cancers and paediatric neuroblastoma, but the challenge has finding ways to effectively target it at effective therapeutic levels. This new research has now moved forward the field sufficiently, such that not only have potential biomarkers of response been identified, but inhibitors are also in advanced preclinical development. This is an exciting new avenue of research that is well worth watching out for in the future.
References:
![]() Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W., Fedorov, O., Morse, E., Keates, T., Hickman, T., Felletar, I., Philpott, M., Munro, S., McKeown, M., Wang, Y., Christie, A., West, N., Cameron, M., Schwartz, B., Heightman, T., La Thangue, N., French, C., Wiest, O., Kung, A., Knapp, S., & Bradner, J. (2010). Selective inhibition of BET bromodomains Nature, 468 (7327), 1067–1073 DOI: 10.1038/nature09504
Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W., Fedorov, O., Morse, E., Keates, T., Hickman, T., Felletar, I., Philpott, M., Munro, S., McKeown, M., Wang, Y., Christie, A., West, N., Cameron, M., Schwartz, B., Heightman, T., La Thangue, N., French, C., Wiest, O., Kung, A., Knapp, S., & Bradner, J. (2010). Selective inhibition of BET bromodomains Nature, 468 (7327), 1067–1073 DOI: 10.1038/nature09504
Puissant, A., Frumm, S., Alexe, G., Bassil, C., Qi, J., Chanthery, Y., Nekritz, E., Zeid, R., Gustafson, W., Greninger, P., Garnett, M., McDermott, U., Benes, C., Kung, A., Weiss, W., Bradner, J., & Stegmaier, K. (2013). Targeting MYCN in Neuroblastoma by BET Bromodomain Inhibition Cancer Discovery, 3 (3), 308–323 DOI: 10.1158/2159–8290.CD–12–0418
Lin, C., Lovén, J., Rahl, P., Paranal, R., Burge, C., Bradner, J., Lee, T., & Young, R. (2012). Transcriptional Amplification in Tumor Cells with Elevated c-Myc Cell, 151 (1), 56–67 DOI: 10.1016/j.cell.2012.08.026
Mertz, J., Conery, A., Bryant, B., Sandy, P., Balasubramanian, S., Mele, D., Bergeron, L., & Sims, R. (2011). Targeting MYC dependence in cancer by inhibiting BET bromodomains Proceedings of the National Academy of Sciences, 108 (40), 16669–16674 DOI: 10.1073/pnas.1108190108
Schnepp, R., & Maris, J. (2013). Targeting MYCN: A Good BET for Improving Neuroblastoma Therapy? Cancer Discovery, 3 (3), 255–257 DOI: 10.1158/2159–8290.CD–13–0018


 Following on from my preview of the
Following on from my preview of the