miRNA primer and resources
Following this morning’s blog post, I’ve been deluged with emails wanting to know more about the basics of miRNA and it’s role in cancer. It’s a very complex topic, as Walter Jessen, a cancer researcher pointed out on Twitter in a discussion with Angela Alexander, a Ph.D student at MD Anderson and myself about miRNA:
“Complicated stuff, esp when expression of mirs doesn’t jive with expression of genes that reg them or that they reg.”
Where reg is ‘regulate’ since Twitter only allows 140 chars or less for communicating ideas between people 🙂 Still, this was exactly the problem I was experiencing when researching the topic recently.
Walter kindly offered the link to a primer he wrote on his excellent blog, Highlight Health, which has some great cancer resources and information. Do check it out.
For those of you already familiar with the technical research in this area, there is a nice blog site specialising in miRNA that is well worth checking out for resources and information.
Memorial Sloan Kettering Cancer Centre in Manhattan have a useful online search tool, which looks at “Predicted microRNA targets & target downregulation scores. Experimentally observed expression patterns.” Be warned, you can become distractible and lose yourself for a hour or two playing with this one 🙂
The University of Southampton in the UK also have a Junk RNA blog site collating some information as well.
Wikipedia has some notes on the topic and some useful diagrams like this one:
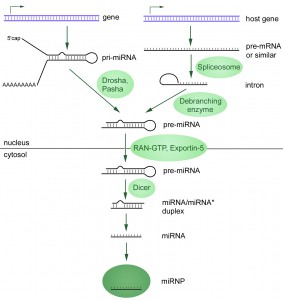 Source: Wikipedia
Source: Wikipedia
Please feel free to add any other useful resources for others to browse in the comments section below.